Zoë Ashwood
PhD student in Computer Science at Princeton University

Email: zashwood at cs dot princeton dot edu
I am a PhD student in the Computer Science Department at Princeton University advised by Jonathan Pillow. During my time at Princeton, my projects have focussed on using machine learning to characterize the behavior of black-box systems, such as animals making decisions. I am interested in exploring the connection between animal and machine learning, and am excited about curriculum learning and automatic curriculum generation.
Keywords: Machine Learning, Reinforcement Learning, Computational Neuroscience and Cognitive Science
Experience
I completed my undergraduate degree (MPhys) in Mathematics and Theoretical Physics at the University of St Andrews in Scotland. Prior to coming to Princeton, I studied as a Robert T. Jones scholar at Emory University and I worked for two years as a Research Fellow for Professor Daniel Ho at Stanford University. At Stanford, we were interested in assessing policy efficacy by carefully designing Randomized-Controlled Trials and applying appropriate statistical methods to measure a policy’s effect.
Selected Projects
Inferring learning rules from animal decision-making
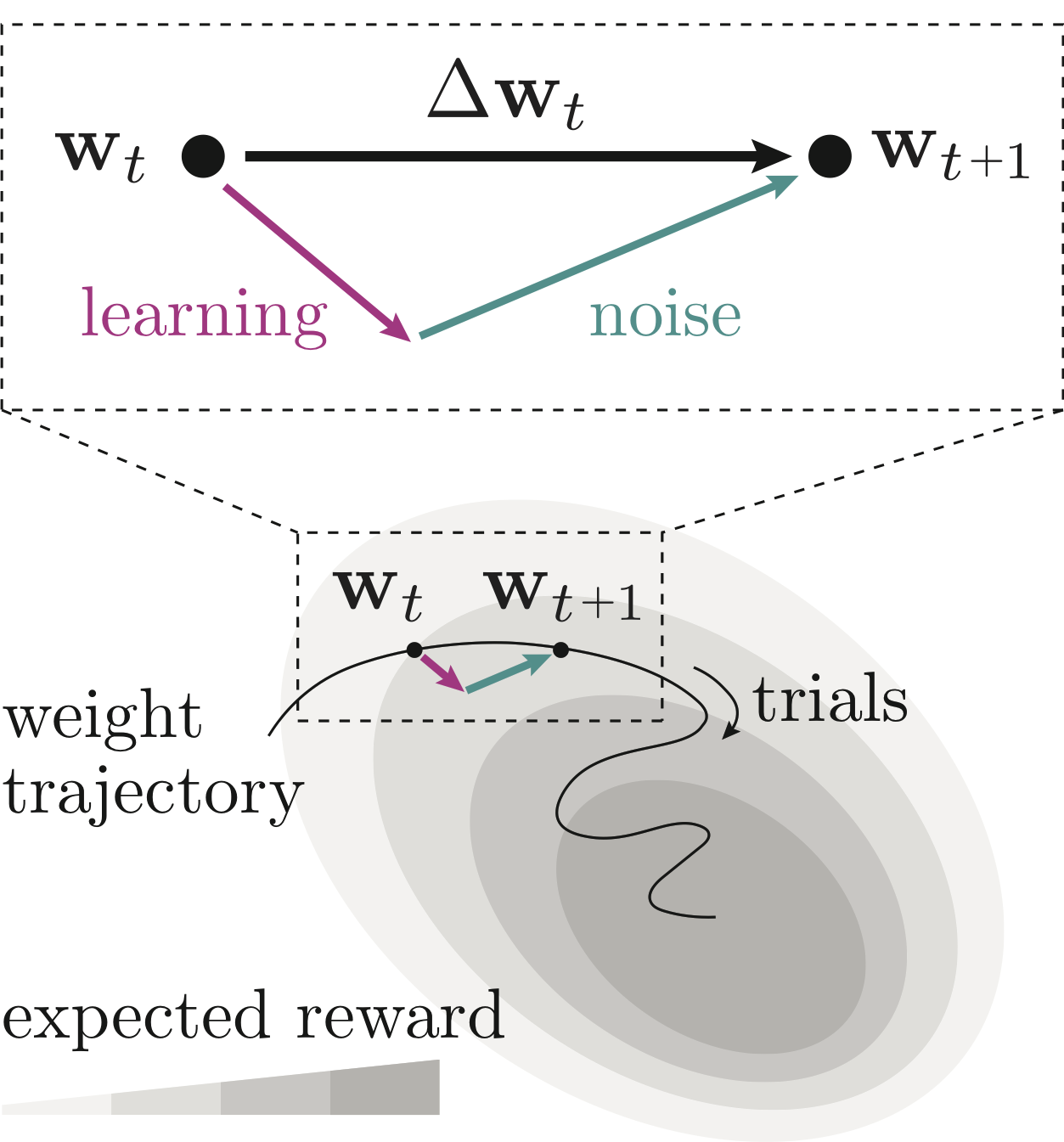 In this project, I worked with Nick Roy, Ji Hyun Bak, the International Brain Laboratory and Jonathan Pillow to develop a model to characterize the behavior of mice and rats learning to perform perceptual decision-making tasks. Our model tracked trial-to-trial changes in the animals’ choice policies, and separated changes into components explainable by a Reinforcement Learning rule, and components that remained unexplained. While the standard REINFORCE learning rule was only able to explain 30% of animals’ trial-to-trial policy updates, REINFORCE with baseline was capable of explaining 92% of updates used by mice learning the “IBL” task. Understanding the rules that underpin animal learning not only provides neuroscientists with insight into their animals, but
also provides concrete examples of biological learning algorithms to the machine
learning community. The paper associated with this project was accepted to NeurIPS 2020, and is linked to above.
In this project, I worked with Nick Roy, Ji Hyun Bak, the International Brain Laboratory and Jonathan Pillow to develop a model to characterize the behavior of mice and rats learning to perform perceptual decision-making tasks. Our model tracked trial-to-trial changes in the animals’ choice policies, and separated changes into components explainable by a Reinforcement Learning rule, and components that remained unexplained. While the standard REINFORCE learning rule was only able to explain 30% of animals’ trial-to-trial policy updates, REINFORCE with baseline was capable of explaining 92% of updates used by mice learning the “IBL” task. Understanding the rules that underpin animal learning not only provides neuroscientists with insight into their animals, but
also provides concrete examples of biological learning algorithms to the machine
learning community. The paper associated with this project was accepted to NeurIPS 2020, and is linked to above.
Mice alternate between discrete strategies during perceptual decision-making
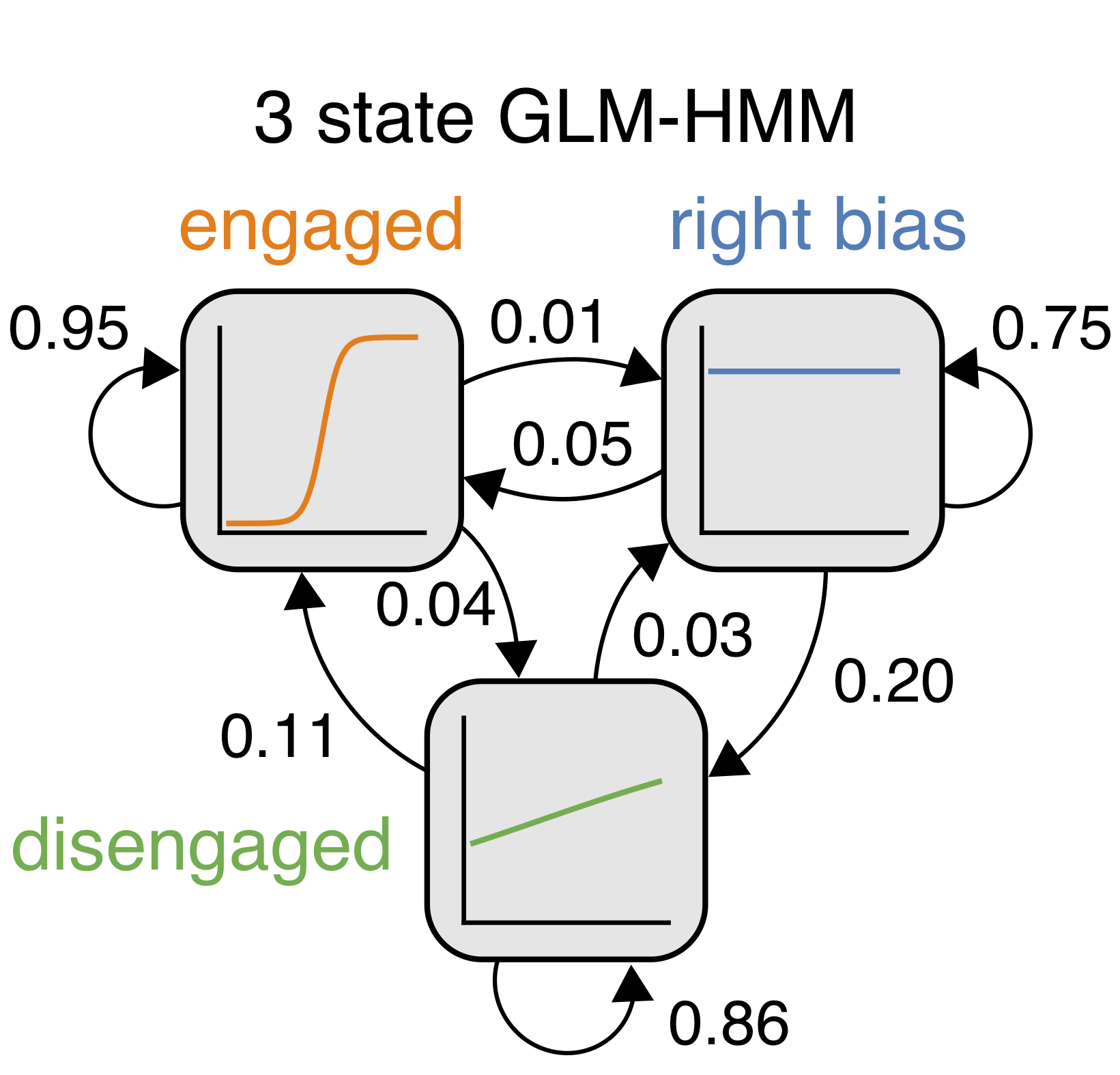
For this project, I worked with Nick Roy, Iris Stone, the International Brain Laboratory, Anne Churchland, Alex Pouget and Jonathan Pillow to implement an Input-Output HMM (IO-HMM) and apply it to mouse decision-making data. We found that discrete latent states underpin mouse choice behavior: with the IO-HMM, we achieved a 5% increase in predictive accuracy of animals’ choices (baseline accuracy was already ∼70%). We presented this work at Cosyne 2020.
Teaching Experience
- 2020 and 2021: Teaching Assistant for Cosyne conference tutorials on “Normative approaches to understanding neural coding and behavior” (2020), as well as tutorial on “Recurrent Neural Networks for Neuroscience” (2021)
- Spring 2019: Assistant Instructor for Princeton’s Neural Networks class (COS 485). Responsibilities were similar to those listed above.
- Fall 2018: Assistant Instructor for Princeton’s Computer Vision (COS 429) class. Responsibilities included holding office hours, answering students’ questions and writing questions for exams.
Leadership, Service and Outreach
- Instructor at Princeton’s first AI4ALL summer camp for rising 11th grade students (2018). AI4ALL is an nonprofit organization devoted to increasing the participation of women and minorities in the field of Artificial Intelligence. For the camp, along with two others, I created a curriculum to introduce the students to NLP. By the end of the summer, our students had replicated one of the leading solutions to the Fake News Challenge.
- Reviewer for UAI and NeurIPS
Awards
- University of St Andrews Principal’s Medal (2014)
- University of St Andrews Prize for Best Project in Theoretical Physics (2014)
- University of St Andrews Class Medal for Physics (2011, 2013)